ESM3 跨越5亿年的蛋白质信息
前言
鄙人电脑网络不太好 只能拿colab跑了,下面是colab链接:generate.ipynb
esm3介绍
论文链接
ESM3,一个可以理解蛋白质序列、结构和功能的多模态生成语言模型。ESM3 可以根据不模态的输入生成全新的、功能性的蛋白质,其生成的蛋白质与已知蛋白质的差异相当于模拟超过 5 亿年的自然进化。
- ESM3使用离散的符号 (token) 来表示蛋白质序列、结构和功能,并通过掩码语言模型的训练目标学习这些符号之间的关系。它能接受来自不同模态的复杂提示,并生成满足这些提示的蛋白质。
- 可编程设计 ESM3 可以根据不同的提示生成蛋白质,包括特定结构、二级结构、表面积、功能关键词等。研究发现,它可以生成与自然界蛋白质截然不同的蛋白质,甚至可以创造全新的蛋白质结构。
- 生物学对齐 通过偏好微调,可以提高 ESM3 遵循复杂提示的能力。更大的模型在对齐后表现出更强的解决复杂问题的能力。
- 生成新的荧光蛋白 利用 ESM3,研究人员成功生成了一个名为 esmGFP 的全新绿色荧光蛋白,其与已知荧光蛋白的序列相似度仅为 58%。这种差异程度相当于超过 5 亿年的自然进化。
使用技术 1. 多模态符号化 (Tokenization):
- 序列: 蛋白质序列被简单地符号化为 20 种常见氨基酸加上特殊符号 (开始、结束、掩码、填充、未知等) 。
- 结构: ESM3 使用一个离散自编码器 (VQ-VAE) 将蛋白质三维结构压缩成离散的结构符号。编码器将每个氨基酸周围的局部结构信息编码为一个符号,解码器则可以将这些符号重建成完整的原子结构。
- 功能: 功能信息通过将 InterPro 和 Gene Ontology (GO) 的文本描述转换为关键词集合来表示。 然后使用局部敏感哈希 (LSH) 将这些关键词量化为离散的符号。
2. 模型架构:
- ESM3 的核心是一个双向 Transformer 网络,它将所有模态的符号融合到一个单一的潜在空间中。
- 为了处理结构信息,ESM3 在第一个 Transformer 块中加入了一个几何注意力 (Geometric Attention) 层,该层可以直接处理原子坐标,捕捉蛋白质三维结构中的几何关系。
3. 训练目标:
- ESM3 使用生成式掩码语言模型的训练目标。在训练过程中,模型会随机掩盖输入符号的一部分,并尝试预测被掩盖的符号。这种训练方式让模型能够学习不同模态符号之间的复杂关系,并生成满足多种约束条件的蛋白质。
4. 生成新蛋白质的算法:
- ESM3 可以根据用户提供的提示生成蛋白质,提示可以是任意模态的组合,例如部分结构、二级结构、功能关键词等。
- 生成过程是迭代进行的,从一个完全掩码的符号序列开始,模型逐步预测每个位置的符号,直到所有符号都被解码出来。
- 在解码过程中,可以使用不同的策略选择下一个要解码的位置,例如熵解码 (entropy decoding) 或最大对数解码 (max logit decoding) 。
- 为了生成高质量的蛋白质,ESM3 还使用了一种联合序列结构优化 (joint sequence structure optimization) 算法,该算法迭代地优化蛋白质序列和结构,以确保生成的蛋白质既满足提示条件,又具有稳定的结构。
5. 生成新的荧光蛋白的例子:
- 研究人员使用 ESM3 生成新的荧光蛋白,首先定义了一个包含关键残基和结构信息的提示,然后使用联合序列结构优化算法迭代地生成和优化候选蛋白质。
- 在生成过程中,研究人员使用各种指标来评估候选蛋白质的质量,例如与模板结构的相似度、序列复杂度、结构稳定性等。
- 通过多次迭代和筛选,研究人员最终生成了一个名为 esmGFP 的全新绿色荧光蛋白,其与已知荧光蛋白的序列差异程度相当于超过 5 亿年的自然进化。
训练方法
库文件导入
1
2
3
4
5
6
7
8
9
10
11
12
13
14
%set_env TOKENIZERS_PARALLELISM=false
!pip install esm
import numpy as np
import torch
!pip install py3Dmol
import py3Dmol
from huggingface_hub import login
from esm.utils.structure.protein_chain import ProteinChain
from esm.models.esm3 import ESM3
from esm.sdk.api import (
ESMProtein,
GenerationConfig,
)
输出如下
在 GPU 上加载 esm-open-small
下面的token需要你自己去注册huggingface的账号并且拿到对应的许可证才行,我自己的token可以用,但是不便于展示
1
2
login(token="********",add_to_git_credential=True)
model = ESM3.from_pretrained("esm3_sm_open_v1", device=torch.device("cuda"))
为 ESM3 构建一个提示,重点关注从天然蛋白质构建基序的任务
使用 esm sdk 中的 ProteinChain 类从 PDB 中获取蛋白质结构
ProteinChain 类是一个可以轻松处理蛋白质结构的对象。它包含 sequence 属性,其中包含蛋白质的氨基酸序列
ProteinChain 还包含一个 atom37_positions numpy 数组,其中包含蛋白质中每个残基的原子坐标
1
2
3
4
5
6
7
8
pdb_id = "1ITU" # PDB ID corresponding to Renal Dipeptidase
chain_id = "A" # Chain ID corresponding to Renal Dipeptidase in the PDB structure
renal_dipep_chain = ProteinChain.from_rcsb(pdb_id, chain_id)
# Alternatively, we could have used ProteinChain.from_pdb() to load a protein structure from a local PDB file
print(renal_dipep_chain.sequence)
print("atom37_positions shape: ", renal_dipep_chain.atom37_positions.shape)
print(renal_dipep_chain.atom37_positions[:3])
数组的形状为 (n_residues, 37, 3) ,其中 n_residues 是蛋白质中残基的数量,37 是可能存在于所有氨基酸中的可能不同原子的数量(例如第一个三个原子是对应于蛋白质主链的 N、C-α 和 C 原子)。 3 对应于每个原子的 x、y 和 z 坐标。蛋白质结构的atom37表示允许我们使用单一格式方便地表示所有氨基酸——坐标仅存在于氨基酸中存在的原子,否则 nan 。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
atom37_positions shape: (369, 37, 3)
[[[-40.525 -9.87 -2.643]
[-39.79 -9.325 -3.825]
[-38.765 -10.354 -4.294]
[-39.096 -8.012 -3.45 ]
[-37.878 -10.748 -3.53 ]
[-38.41 -7.359 -4.629]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-39.105 -7.036 -5.617]
[-37.177 -7.161 -4.562]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]]
[[-38.877 -10.768 -5.555]
[-37.975 -11.767 -6.115]
[-36.508 -11.389 -6.096]
[-38.365 -12.141 -7.546]
[-35.674 -12.205 -5.716]
[-37.411 -13.109 -8.19 ]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-36.568 -12.698 -9.215]
[-37.342 -14.432 -7.756]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-35.67 -13.589 -9.799]
[-36.447 -15.332 -8.333]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-35.612 -14.91 -9.356]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]]
[[-36.191 -10.172 -6.525]
[-34.798 -9.736 -6.576]
[-34.127 -9.485 -5.225]
[-34.629 -8.57 -7.553]
[-32.912 -9.65 -5.097]
[-34.691 -8.997 -9.002]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-33.837 -9.991 -9.482]
[-35.629 -8.45 -9.87 ]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-33.912 -10.442 -10.806]
[-35.714 -8.891 -11.195]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[-34.852 -9.892 -11.662]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]
[ nan nan nan]]]
蛋白质预测可视化
1
2
3
4
5
6
7
8
9
10
11
12
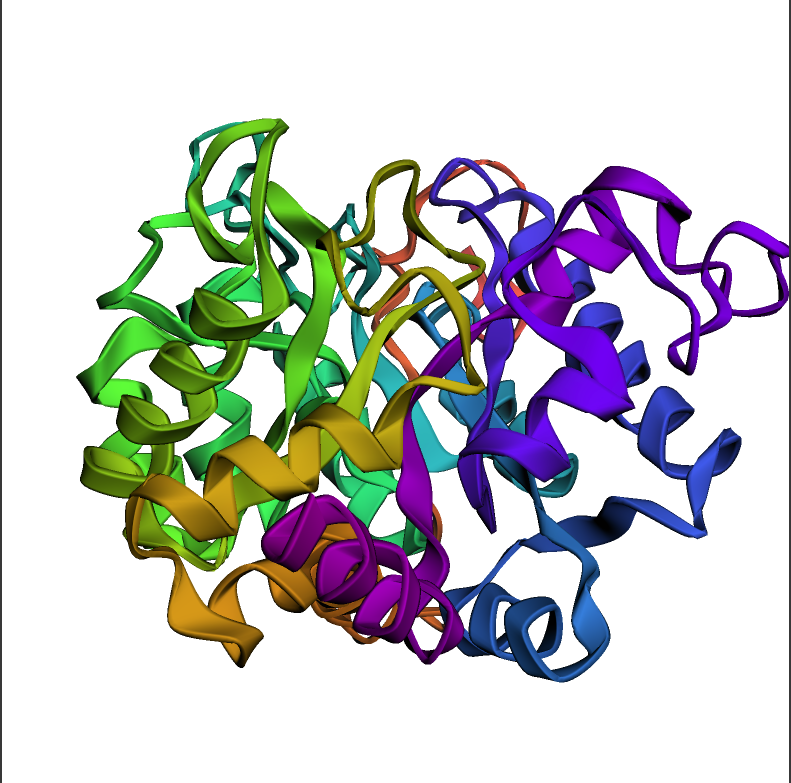
# First we can create a `py3Dmol` view object
view = py3Dmol.view(width=500, height=500)
# py3Dmol requires the atomic coordinates to be in PDB format, so we convert the `ProteinChain` object to a PDB string
pdb_str = renal_dipep_chain.to_pdb_string()
# Load the PDB string into the `py3Dmol` view object
view.addModel(pdb_str, "pdb")
# Set the style of the protein chain
view.setStyle({"cartoon": {"color": "spectrum"}})
# Zoom in on the protein chain
view.zoomTo()
# Display the protein chain
view.show()
这个实际上是可以3维旋转的 十分震撼
使用 ESM3 构建该蛋白质的基序
我们将使用肾二肽酶的螺旋线圈基序的序列和结构提示模型,并让模型生成包含该基序的更大支架
1
2
3
4
5
6
motif_inds = np.arange(123, 146)
# `ProteinChain` objects can be indexed like numpy arrays to extract the sequence and atomic coordinates of a subset of residues
motif_sequence = renal_dipep_chain[motif_inds].sequence
motif_atom37_positions = renal_dipep_chain[motif_inds].atom37_positions
print("Motif sequence: ", motif_sequence)
print("Motif atom37_positions shape: ", motif_atom37_positions.shape)
输出如下
1
2
Motif sequence: VEGGHSIDSSLGVLRALYQLGMR
Motif atom37_positions shape: (23, 37, 3)
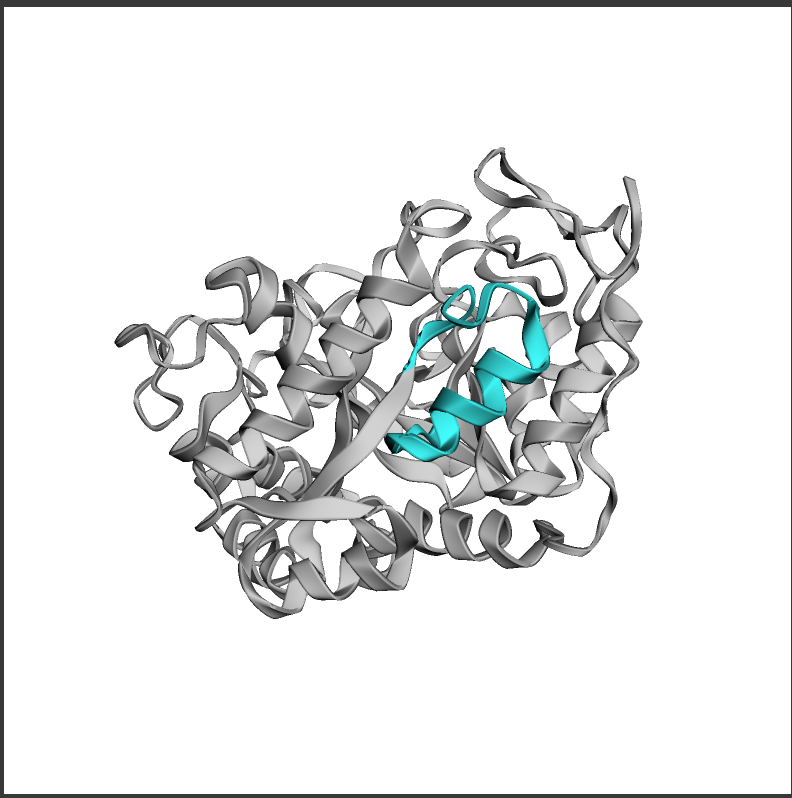
使用 py3Dmol 可视化原始链中的主题。我们将原始链条涂成灰色,图案涂成蓝色
1
2
3
4
5
6
7
view = py3Dmol.view(width=500, height=500)
view.addModel(pdb_str, "pdb")
view.setStyle({"cartoon": {"color": "lightgrey"}})
motif_res_inds = (motif_inds + 1).tolist() # residue indices are 1-indexed in PDB files, so we add 1 to the indices
view.addStyle({"resi": motif_res_inds}, {"cartoon": {"color": "cyan"}})
view.zoomTo()
view.show()
使用 ESMProtein 类构建一个提示,指示 ESM3 构建主题
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
prompt_length = 200
# First, we can construct a sequence prompt of all masks
sequence_prompt = ["_"]*prompt_length
# Then, we can randomly insert the motif sequence into the prompt (we randomly choose 72 here)
sequence_prompt[72:72+len(motif_sequence)] = list(motif_sequence)
sequence_prompt = "".join(sequence_prompt)
print("Sequence prompt: ", sequence_prompt)
print("Length of sequence prompt: ", len(sequence_prompt))
# Next, we can construct a structure prompt of all nan coordinates
structure_prompt = torch.full((prompt_length, 37, 3), np.nan)
# Then, we can insert the motif atomic coordinates into the prompt, starting at index 72
structure_prompt[72:72+len(motif_atom37_positions)] = torch.tensor(motif_atom37_positions)
print("Structure prompt shape: ", structure_prompt.shape)
print("Indices with structure conditioning: ", torch.where(~torch.isnan(structure_prompt).any(dim=-1).all(dim=-1))[0].tolist())
# Finally, we can use the ESMProtein class to compose the sequence and structure prompts into a single prompt that can be passed to ESM3
protein_prompt = ESMProtein(sequence=sequence_prompt, coordinates=structure_prompt)
输出
1
2
3
4
Sequence prompt: ________________________________________________________________________VEGGHSIDSSLGVLRALYQLGMR_________________________________________________________________________________________________________
Length of sequence prompt: 200
Structure prompt shape: torch.Size([200, 37, 3])
Indices with structure conditioning: [72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94]
使用模型的 generate 方法根据提示对蛋白质序列进行迭代采样。在底层,模型执行 num_steps 前向传递,并在每一步对一组标记进行采样,直到所生成的所选轨道完全未被屏蔽。
1
2
3
4
5
6
7
8
9
10
11
# We'll have to first construct a `GenerationConfig` object that specifies the decoding parameters that we want to use
sequence_generation_config = GenerationConfig(
track="sequence", # We want ESM3 to generate tokens for the sequence track
num_steps=sequence_prompt.count("_") // 2, # We'll use num(mask tokens) // 2 steps to decode the sequence
temperature=0.5, # We'll use a temperature of 0.5 to control the randomness of the decoding process
)
# Now, we can use the `generate` method of the model to decode the sequence
sequence_generation = model.generate(protein_prompt, sequence_generation_config)
print("Sequence Prompt:\n\t", protein_prompt.sequence)
print("Generated sequence:\n\t", sequence_generation.sequence)
输出
1
2
3
4
5
100%|██████████| 88/88 [00:20<00:00, 4.26it/s]
Sequence Prompt:
________________________________________________________________________VEGGHSIDSSLGVLRALYQLGMR_________________________________________________________________________________________________________
Generated sequence:
LERLREGGVGAQFWSVYVPAEYKGEEAVRKTLEQIDLVHRTVERHPDKFELATTAADIDRIKAQGKIASLIGVEGGHSIDSSLGVLRALYQLGMRYMTLTWNKNLPWADSATDEHKDHNGLSDFGKEVVREMNRLGMLVDVSHVSEDTFWDALEVSTAPIIASHSSARALTDHPRNMTDEQLRALKENGGVMMVNFYPE
使用 generate 方法通过迭代采样结构标记来预测生成序列的结构,并且使用 py3Dmol 可视化生成的结构。我们将可视化生成的结构(右,绿色)以及绘制图案的原始结构(左,灰色)。图案残基呈青色。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
structure_prediction_config = GenerationConfig(
track="structure", # We want ESM3 to generate tokens for the structure track
num_steps=len(sequence_generation) // 8,
temperature=0.7,
)
structure_prediction_prompt = ESMProtein(sequence=sequence_generation.sequence)
structure_prediction = model.generate(structure_prediction_prompt, structure_prediction_config)
# Convert the generated structure to a back into a ProteinChain object
structure_prediction_chain = structure_prediction.to_protein_chain()
# Align the generated structure to the original structure using the motif residues
motif_inds_in_generation = np.arange(72, 72+len(motif_sequence))
structure_prediction_chain.align(renal_dipep_chain, mobile_inds=motif_inds_in_generation, target_inds=motif_inds)
crmsd = structure_prediction_chain.rmsd(renal_dipep_chain, mobile_inds=motif_inds_in_generation, target_inds=motif_inds)
print("cRMSD of the motif in the generated structure vs the original structure: ", crmsd)
view = py3Dmol.view(width=1000, height=500, viewergrid=(1, 2))
view.addModel(pdb_str, "pdb", viewer=(0, 0))
view.addModel(structure_prediction_chain.to_pdb_string(), "pdb", viewer=(0, 1))
view.setStyle({"cartoon": {"color": "lightgrey"}}, viewer=(0, 0))
view.setStyle({"cartoon": {"color": "lightgreen"}}, viewer=(0, 1))
view.addStyle({"resi": motif_res_inds}, {"cartoon": {"color": "cyan"}}, viewer=(0, 0))
view.addStyle({"resi": (motif_inds_in_generation+1).tolist()}, {"cartoon": {"color": "cyan"}}, viewer=(0, 1))
view.zoomTo()
view.show()
输出如下
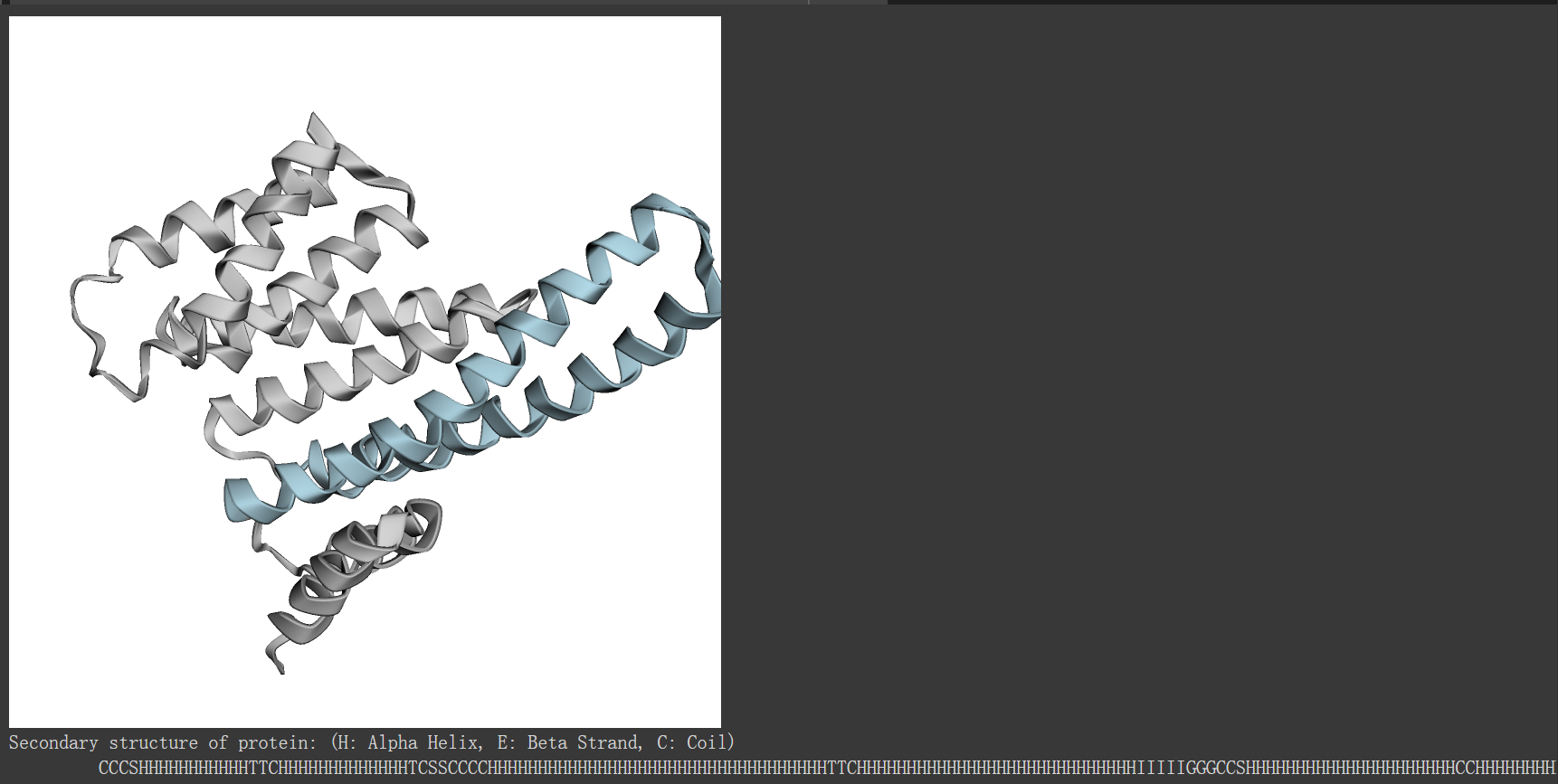
二级结构编辑示例:螺旋缩短
1
2
3
4
5
6
7
8
9
10
helix_shortening_chain = ProteinChain.from_rcsb("7XBQ", "A")
view = py3Dmol.view(width=500, height=500)
view.addModel(helix_shortening_chain.to_pdb_string(), "pdb")
view.setStyle({"cartoon": {"color": "lightgrey"}})
helix_region = np.arange(38, 111) # zero-indexed
view.addStyle({"resi": (helix_region + 1).tolist()}, {"cartoon": {"color":"lightblue"}})
view.zoomTo()
view.show()
helix_shortening_ss8 = "CCCSHHHHHHHHHHHTTCHHHHHHHHHHHHHTCSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHTTCHHHHHHHHHHHHHHHHHHHHHHHHHHHHIIIIIGGGCCSHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHSCTTCHHHHHHHHHHHHHIIIIICCHHHHHHHHHHHHHHHHTTCTTCCSSHHHHHHHHHHHHHHHHHHHC"
print("Secondary structure of protein: (H: Alpha Helix, E: Beta Strand, C: Coil) \n\t", helix_shortening_ss8)
原始蛋白质中的螺旋-螺旋-螺旋区域有 73 个残基长。我们将通过使用部分序列和二级结构提示模型来尝试将其缩短至45个残基
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
shortened_region_length = 45
# We'll construct a sequence prompt that masks the (shortened) helix-coil-helix region, but leaves the flanking regions unmasked
sequence_prompt = helix_shortening_chain.sequence[:helix_region[0]] + "_" * shortened_region_length + helix_shortening_chain.sequence[helix_region[-1] + 1:]
print("Sequence prompt:\n\t", sequence_prompt)
# We'll construct a secondary structure prompt that retains the secondary structure of the flanking regions, and shortens the lengths of helices in the helix-coil-helix region
ss8_prompt = helix_shortening_ss8[:helix_region[0]] + (((shortened_region_length - 3) // 2) * "H" + "C"*3 + ((shortened_region_length - 3) // 2) * "H") + helix_shortening_ss8[helix_region[-1] + 1:]
print("SS8 prompt:\n\t", ss8_prompt)
print("Proposed SS8 for shortened helix-coil-helix region:\n\t", " "*helix_region[0] + ss8_prompt[helix_region[0]:helix_region[0]+45])
print("")
print("Original sequence:\n\t", helix_shortening_chain.sequence)
print("Original SS8:\n\t", helix_shortening_ss8)
print("Original SS8 for helix-coil-helix region:\n\t", " "*helix_region[0] + helix_shortening_ss8[helix_region[0]:helix_region[-1]+1])
# We can again use the ESMProtein class to compose the sequence and secondary structure prompts into a single prompt that can be passed to ESM3
protein_prompt = ESMProtein(sequence=sequence_prompt, secondary_structure=ss8_prompt)
输出:
1
2
3
4
5
6
7
8
9
10
11
12
13
Sequence prompt:
MAREENVYMAKLAEQAERYEEMVQFMEKVSTSLGSEEL_____________________________________________SASNGDSKVFYLKMKGDYHRYLAEFKTGAERKEAAESTLSAYKAAQDIANTELAPTHPIRLGLALNFSVFYYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWT
SS8 prompt:
CCCSHHHHHHHHHHHTTCHHHHHHHHHHHHHTCSSCCCHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHGCCSHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHSCTTCHHHHHHHHHHHHHIIIIICCHHHHHHHHHHHHHHHHTTCTTCCSSHHHHHHHHHHHHHHHHHHHC
Proposed SS8 for shortened helix-coil-helix region:
HHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHH
Original sequence:
MAREENVYMAKLAEQAERYEEMVQFMEKVSTSLGSEELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEESRGNEEHVKCIKEYRSKIESELSNICDGILKLLDSNLIPSASNGDSKVFYLKMKGDYHRYLAEFKTGAERKEAAESTLSAYKAAQDIANTELAPTHPIRLGLALNFSVFYYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWT
Original SS8:
CCCSHHHHHHHHHHHTTCHHHHHHHHHHHHHTCSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHTTCHHHHHHHHHHHHHHHHHHHHHHHHHHHHIIIIIGGGCCSHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHSCTTCHHHHHHHHHHHHHIIIIICCHHHHHHHHHHHHHHHHTTCTTCCSSHHHHHHHHHHHHHHHHHHHC
Original SS8 for helix-coil-helix region:
CHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHTTCHHHHHHHHHHHHHHHHHHHHHHHHHHHHIIIIIGG
我们可以再次使用模型的 generate 方法根据提示迭代解码蛋白质序列
1
2
3
4
print("Generating protein sequence...")
sequence_generation = model.generate(protein_prompt, GenerationConfig(track="sequence", num_steps=protein_prompt.sequence.count("_") // 2, temperature=0.5))
print("Folding protein...")
structure_prediction = model.generate(ESMProtein(sequence=sequence_generation.sequence), GenerationConfig(track="structure", num_steps=len(protein_prompt) // 4, temperature=0))
输出:
1
2
3
4
Generating protein sequence...
100%|██████████| 22/22 [00:04<00:00, 4.49it/s]
Folding protein...
100%|██████████| 51/51 [00:11<00:00, 4.51it/s]
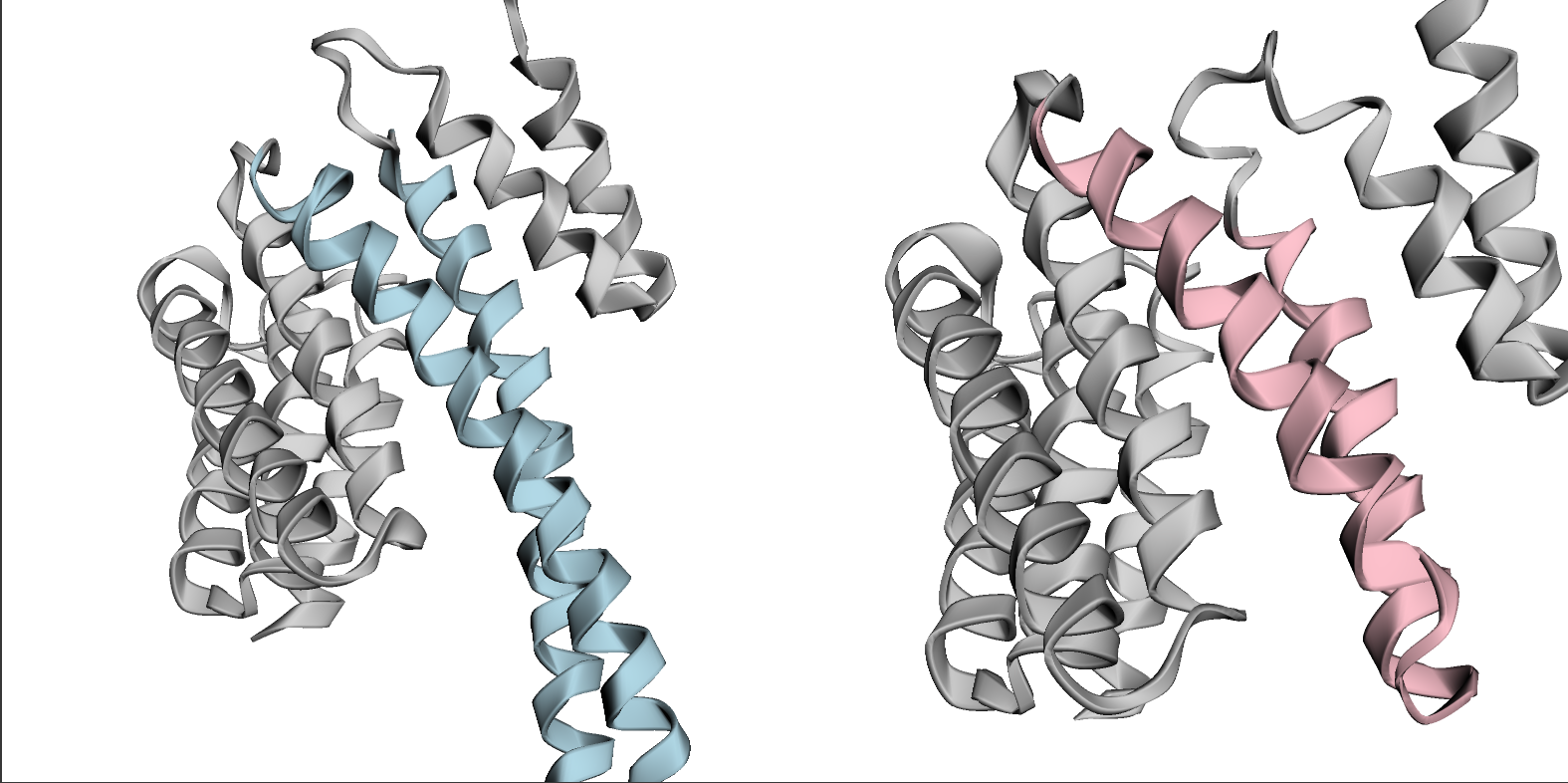
使用 py3Dmol 可视化生成的结构。我们将可视化生成的结构(右)以及绘制主题的原始结构(左)。原始结构中的螺旋-螺旋-螺旋区域为蓝色,生成结构中的缩短区域为粉色。
1
2
3
4
5
6
7
8
9
10
predicted_chain = structure_prediction.to_protein_chain()
predicted_chain = predicted_chain.align(helix_shortening_chain, mobile_inds=np.arange(len(predicted_chain) - 120, len(predicted_chain)), target_inds=np.arange(len(helix_shortening_chain) - 120, len(helix_shortening_chain)))
view = py3Dmol.view(width=1000, height=500, viewergrid=(1, 2))
view.addModel(helix_shortening_chain.to_pdb_string(), "pdb", viewer=(0, 0))
view.addModel(predicted_chain.to_pdb_string(), "pdb", viewer=(0, 1))
view.setStyle({"cartoon": {"color": "lightgrey"}})
view.addStyle({"resi": (helix_region + 1).tolist()}, {"cartoon": {"color":"lightblue"}},viewer=(0, 0))
view.addStyle({"resi": (np.arange(helix_region[0], helix_region[0] + 45) + 1).tolist()}, {"cartoon": {"color":"pink"}},viewer=(0, 1))
view.zoomTo()
view.show()
SASA 编辑示例:暴露隐藏的螺旋
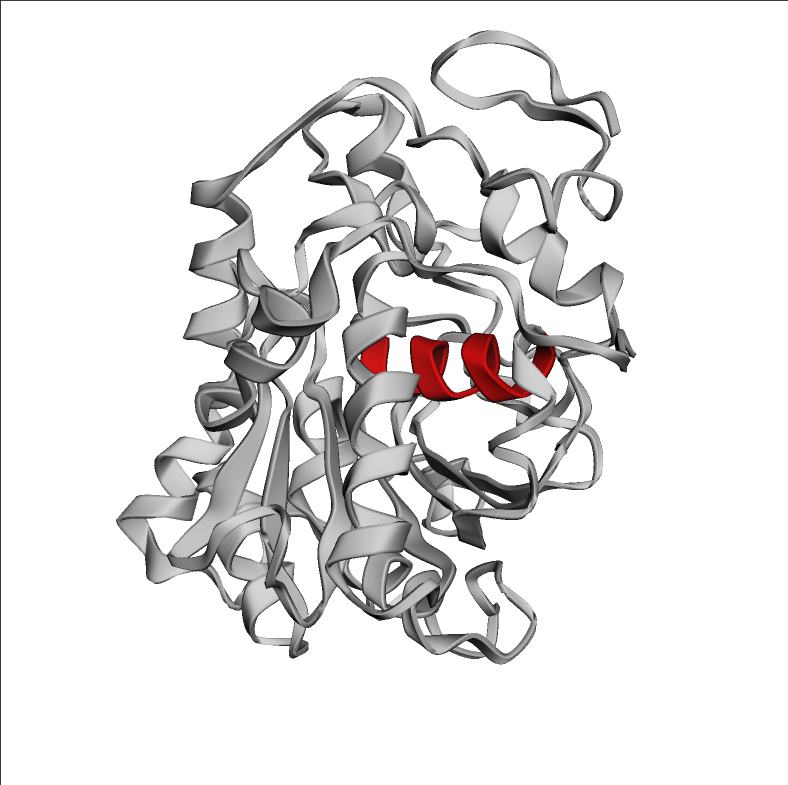
从 PDB 中获取 1LBS 并使用 py3Dmol 将其可视化。 1LBS 有交替的 α-β 夹层折叠,中心有埋入式螺旋,以红色突出显示
1
2
3
4
5
6
7
8
9
10
lipase_chain = ProteinChain.from_rcsb("1LBS", "A")
span_start = 105
span_end = 116
view = py3Dmol.view(width=500, height=500)
view.addModel(lipase_chain.to_pdb_string(), "pdb")
view.setStyle({"cartoon": {"color": "lightgrey"}})
view.addStyle({"resi": (np.arange(span_start, span_end) + 1).tolist()}, {"cartoon": {"color":"red"}})
view.zoomTo()
view.show()
lipase_ss8 = "CCSSCCCCSSCHHHHHHTEEETTBBTTBCSSEEEEECCTTCCHHHHHTTTHHHHHHHTTCEEEEECCTTTTCSCHHHHHHHHHHHHHHHHHHTTSCCEEEEEETHHHHHHHHHHHHCGGGGGTEEEEEEESCCTTCBGGGHHHHHTTCBCHHHHHTBTTCHHHHHHHHTTTTBCSSCEEEEECTTCSSSCCCCSSSTTSTTCCBTSEEEEHHHHHCTTCCCCSHHHHHBHHHHHHHHHHHHCTTSSCCGGGCCSTTCCCSBCTTSCHHHHHHHHSTHHHHHHHHHHSCCBSSCCCCCGGGGGGSTTCEETTEECCC"
为 ESM3 构建一个多模式提示来指示它暴露隐藏的螺旋,如下所示:
提示以红色突出显示的隐藏螺旋的结构 - 这将提示 ESM3 生成包含相同螺旋的蛋白质
以高 SASA 值提示隐藏螺旋中的残基 - 这将促使 ESM3 将螺旋暴露于蛋白质表面
1
2
3
4
5
6
7
8
9
structure_prompt = torch.full((len(lipase_chain), 37, 3), torch.nan)
structure_prompt[span_start:span_end] = torch.tensor(lipase_chain[span_start:span_end].atom37_positions, dtype=torch.float32)
sasa_prompt = [None]*len(lipase_chain)
sasa_prompt[span_start:span_end] = [40.0]*(span_end - span_start)
print("SASA prompt (just for buried region): ", sasa_prompt[span_start:span_end])
protein_prompt = ESMProtein(sequence="_"*len(lipase_chain), coordinates=structure_prompt, sasa=sasa_prompt)
输出
1
SASA prompt (just for buried region): [40.0, 40.0, 40.0, 40.0, 40.0, 40.0, 40.0, 40.0, 40.0, 40.0, 40.0]
在这里抽取 32 个样本,并按 ESM3 预测的 TM 分数 (pTM) 最高的代进行排序。
1
2
3
4
5
6
7
8
9
10
11
generated_proteins = []
N_SAMPLES = 32
for i in range(N_SAMPLES):
print("Generating protein sequence...")
sequence_generation = model.generate(protein_prompt, GenerationConfig(track="sequence", num_steps=len(protein_prompt) // 8, temperature=0.7))
print("Folding protein...")
structure_prediction = model.generate(ESMProtein(sequence=sequence_generation.sequence), GenerationConfig(track="structure", num_steps=len(protein_prompt) // 32))
generated_proteins.append(structure_prediction)
# Sort generations by ptm
generated_proteins = sorted(generated_proteins, key=lambda x: x.ptm.item(), reverse=True)
输出
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
Generating protein sequence...
100%|██████████| 39/39 [00:12<00:00, 3.14it/s]
Folding protein...
100%|██████████| 9/9 [00:02<00:00, 3.02it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:12<00:00, 3.01it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.96it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.95it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.91it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.88it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.80it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.78it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.81it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.85it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.87it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.87it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.85it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.86it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.82it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.81it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.85it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.85it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.86it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.82it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.82it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.81it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.82it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.85it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.85it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.84it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.84it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.82it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.79it/s]
Generating protein sequence...
100%|██████████| 39/39 [00:13<00:00, 2.83it/s]
Folding protein...
100%|██████████| 9/9 [00:03<00:00, 2.83it/s]
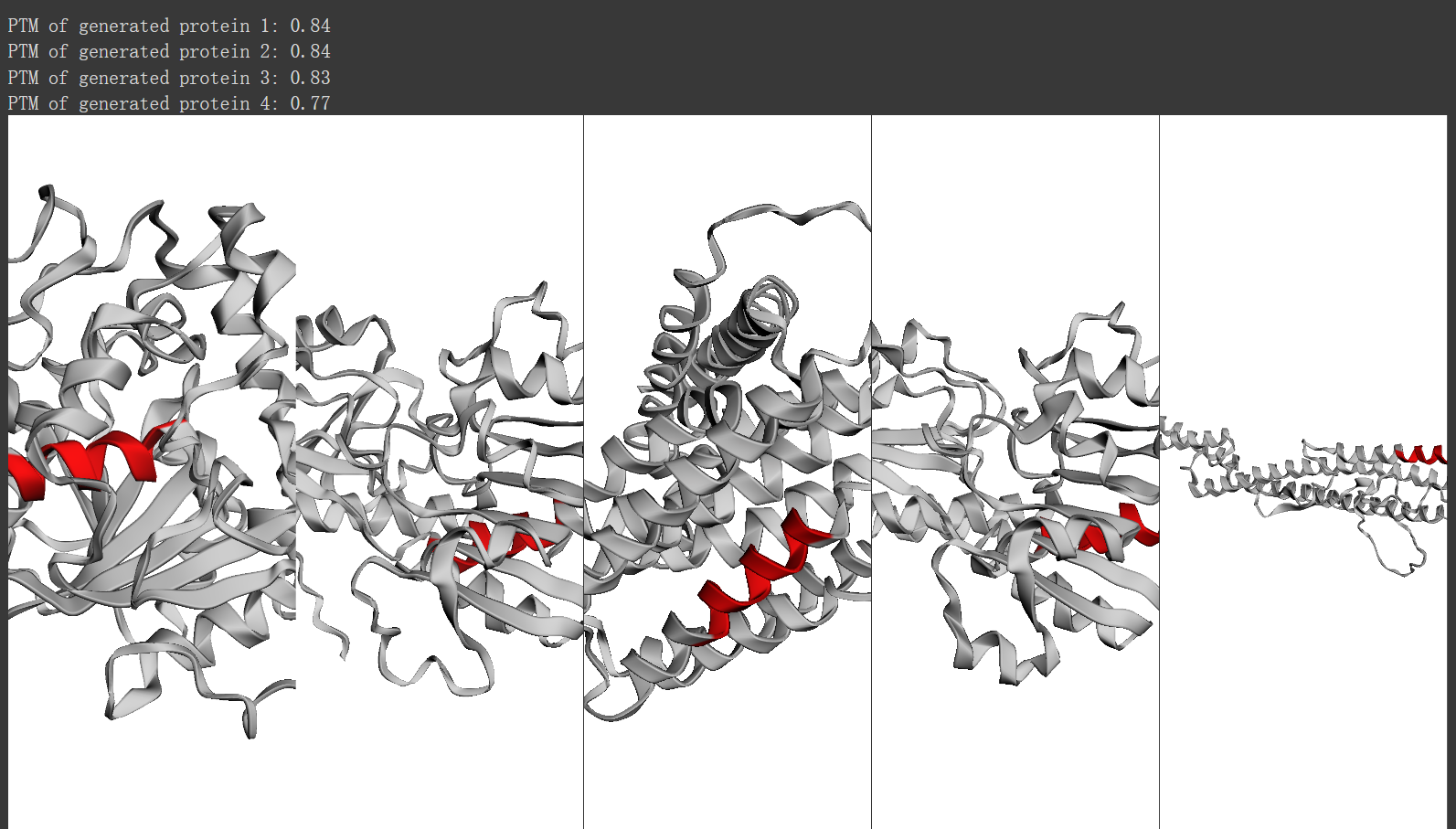
通过 pTM 可视化前 4 代以及原始蛋白质(左侧)
1
2
3
4
5
6
7
8
9
10
N_SAMPLES_TO_SHOW = 4
view = py3Dmol.view(width=1000, height=500, viewergrid=(1, N_SAMPLES_TO_SHOW+1))
view.addModel(lipase_chain.to_pdb_string(), "pdb", viewer=(0, 0))
for i in range(N_SAMPLES_TO_SHOW):
print("PTM of generated protein {}: {:.2f}".format(i+1, generated_proteins[i].ptm.item()))
view.addModel(generated_proteins[i].to_protein_chain().to_pdb_string(), "pdb", viewer=(0, i+1))
view.setStyle({"cartoon": {"color": "lightgrey"}})
view.addStyle({"resi": (np.arange(span_start, span_end) + 1).tolist()}, {"cartoon": {"color": "red"}})
view.zoomTo()
view.show()